




The important membrane protein structures of apoptosome are published on 《Cell》by users of SSRF Macromolecular beamline
The research groups led by Professors Yigong Shi and Nieng Yan at Tsinghua University in Beijing have now determined the protein structure of CED-4 apoptosome and published the results on the journal Cell(”Crystal Structure of the Caenorhabditis elegans Apoptosome Reveals an Octameric Assembly of CED-4”; Volume 141, Issue 3, 30 April 2010, Pages 446-457).
Prof. Yigong Shi’s group has been working on the mechanistic study of the regulation of apoptosis for almost a decade. Prof. Nieng Yan was awarded the Young Scientist Award (North America) by Science/AAAS and GE Healthcare in 2005 for her thesis research on the structural and biochemical characterization of the apoptotic pathway in the nematode C. elegans.
The initiation of programmed cell death in C. elegans is controlled by a linear pathway.

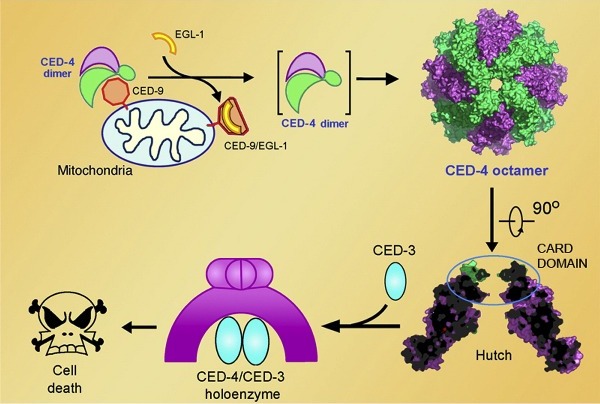
Cell death is executed by CED-3, a representative member of a family of proteases called caspases. CED-3 is synthesized as an inactive zymogen, whose auto-activation requires the adaptor protein CED-4. In normal cells, CED-4 is sequestered by CED-9 (homologous to mammalian Bcl-2) into a (CED-4)2:CED-9 complex, in which the CED-4 molecules exist as an asymmetric dimer. In the cells that are programmed to die, an upstream protein EGL-1 (analogous to mammalian BH3-only proteins) binds to CED-9 and releases the CED-4 dimer, which undergoes further oligomerization to form an active CED-4 apoptosome. In this pathway, a key question remained unclear, that is, how CED-4 activates CED-3. With 10 years struggle, they finally solved the 3.8 angstrom structure of CED-4 in 2008. It took more than 20 synchrotron trips in US, Japan and China to further improve the resolution. Finally, the protein structure was determined at 3.55 angstrom resolution with the data collected at SSRF. They also determined the structure of CED-4:CED-3 complex at 3.8 anstrom.
The CED-4 apoptosome, which comprises eight CED-4 molecules, has a structure that resembles a funnel (see image below). Each CED-4 molecule contains an ATP molecule and an Mg2+ ion — both deeply buried inside the CED-4 molecule — as well as a CARD( caspase-recruiting domain) at the amino terminal. Eight CARDs in turn bundle together to form two staggered rings at the narrow end of the funnel. After incubating CED-4 and CED-3, CED-4:CED-3 complex was obtained. Surprisingly, the molar ratio of CED-4 to CED-3 in this complex is approximately 4:1. On the basis of structure and biochemical characterizations, they proposed a working model for CED-4 mediated CED-3 activation. CED-4 octamer recruits 2 CED-3 molecules into the hutch, promotes the dimerization of CED-3, and enhances CED-3’s protease activity. As an important member of AAA+ family and NB-ARC(nucleotide-binding, Apaf-1,R proteins, and CED-4)family, the structure of CED-4 also reveals some general structural principles of these protein, which sheds light on the study of the structure and function of this large family of proteins.