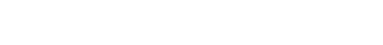





Programming Cell Adhesion for On-Chip Sequential Boolean Logic Functions
Researchers in Shanghai Institute of Applied Physics CAS recently developed a new method to program cell adhesion for on-chip sequential boolean logic functions, which was published on J. Am. Chem. Soc.2017, 139, 10176。
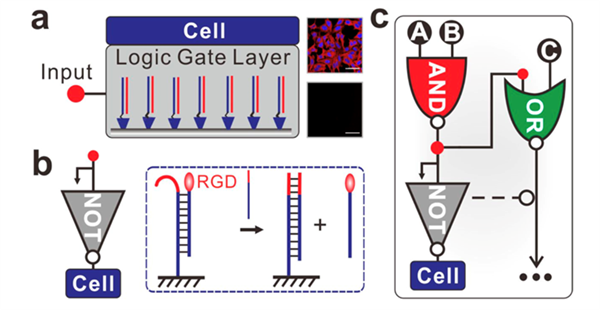
Chemical reaction networks (CRNs) play important roles in various important cellular processes, which allow dynamic regulation of cell–cell communications and interactions between mammalian cells and the extracellular matrix (ECM).In this work, authors developed in vitro constructed DNA-based CRNs to program on-chip cell adhesion. They found that the RGD-functionalized DNA CRNs are entirely noninvasive when interfaced with the fluidic mosaic membrane of living cells. DNA toehold with different lengths could tunably alter the release kinetics of cells, which shows rapid release in minutes with the use of a 6-base toehold. They further demonstrated the realization of Boolean logic functions by using DNA strand displacement reactions, which include multi-input and sequential cell logic gates (AND, OR, XOR, and AND-OR). This study provides a highly generic tool for self-organization of biological systems.